ADCAIJ: Advances in Distributed Computing and Artificial Intelligence Journal
Regular Issue, Vol. 13 (2024), e31762
eISSN: 2255-2863
DOI: https://doi.org/10.14201/adcaij.31762
CyberUnits Bricks: An Implementation Study of a Class Library for Simulating Nonlinear Biological Feedback Loops
Johannes W. Dietricha,b,c,d,e, Nina Siegmara,b,c,d,e, Jonas R. Hojjatia,b,c,d,e, Oliver Gardtf and Bernhard O. Boehmg,h
a Diabetes, Endocrinology and Metabolism Section, Department of Medicine I, St. Josef University Hospital, UK RUB, Gudrunstr. 56, D-44791 Bochum, Germany. johannes.dietrich@ruhr-uni-bochum.de
b Diabetes Centre Bochum/Hattingen, Blankenstein Hospital, Im Vogelsang 5–11, D-45527 Hattingen, Germany
c Centre for Diabetes Technology (ZDT), Catholic Hospitals Bochum, Gudrunstr. 56, D-44791 Bochum, Germany
d Centre for Rare Endocrine Diseases (ZSEK), Ruhr Center for Rare Diseases (CeSER), Ruhr University of Bochum and Witten/Herdecke University, Alexandrinenstr. 5, D-44791 Bochum, Germany
e Centre for Thyroid Medicine (ZSM), Catholic Hospitals Bochum, Gudrunstr. 56, D-44791 Bochum, Germany
f Rheumatology Section, Department of Medicine I, Blankenstein Hospital, Im Vogelsang 5–11, D-45527 Hattingen, Germany
g Lee Kong Chian School of Medicine, Nanyang Technological University Singapore, 11 Mandalay Road, Singapore 308232, Singapore
h King’s College London, School of Life Course & Population Sciences, London U.K.
ABSTRACT
Feedback loops and other types of information processing structures play a pivotal role in maintaining the internal milieu of living organisms. Although methods of biomedical cybernetics and systems biology help to translate between the structure and function of processing structures, computer simulations are necessary for studying nonlinear systems and the full range of dynamic responses of feedback control systems.
Currently, available approaches for modelling and simulation comprise basically domain-specific environments, toolkits for computer algebra systems and custom software written in universal programming languages for a specific purpose, respectively. All of these approaches are faced with certain weaknesses. We therefore developed a cross-platform class library that provides versatile building bricks for writing computer simulations in a universal programming language (CyberUnits Bricks). It supports the definition of models, the simulative analysis of linear and nonlinear systems in the time and frequency domain and the plotting of block diagrams.
We compared several programming languages that are commonly used in biomedical research (S in the R implementation and Python) or that are optimized for speed (Swift, C++ and Object Pascal). In benchmarking experiments with two prototypical feedback loops, we found the implementations in Object Pascal to deliver the fastest results.
CyberUnits Bricks is available as open-source software that has been optimised for Embarcadero Delphi and the Lazarus IDE for Free Pascal.
KEYWORDS
Modelling and simulation; Systems biology; Object Pascal; Nonlinear systems; Computational biomedicine.
1. Introduction
In an unpredictable and commonly adverse environment, the survival of organisms depends on mechanisms constantly controlling their internal milieu. These processes render key physiological properties constant (homeostasis), adaptive (type 1 allostasis) or predictive (type 2 allostasis) (McEwen and Stellar, 1993; McEwen, 1998; Schulkin and Sterling, 2019; Hoermann et al., 2022). Disturbances of homeostatic processes as well as impaired or excessive allostatic responses belong to the major causes of acute and chronic disease. Therefore, insights into the function of biological feedforward and feedback circuits that underlie homeostatic and allostatic response patterns help to understand the evolution of health and disease, and they provide advanced methods for diagnostic investigation and therapy planning (Dietrich et al., 2016; Han et al., 2016; Alon, 2020; Cruz-Loya et al., 2022; Dietrich et al., 2022). Several related transdisciplinary fields such as biomedical cybernetics, systems biology and systems medicine address the translation between the structure and function of control motifs (Tretter, 2018; Tretter et al., 2021; Head et al., 2022). Additionally, characteristic dynamical patterns or shapes that emerge from this kind of structure-function coupling, including oscillation, multistability and chaotic behaviour, have been demonstrated (Alon, 2007; Kolar-Anic et al., 2023; Tretter et al., 2023). However, the nonlinear nature of most biological information processing structures sets limits to their mathematical analysis. Computer simulations help close this gap. Additionally, they provide insights into the temporal evolution of system states and support methods of sensitivity analysis (Curcio et al., 2020; Dietrich, 2017; Dietrich and Boehm, 2021; Pompa et al., 2021).
1.1. State of the Art
Modelling and simulation in life sciences is challenged by the fact that traditional approaches of control theory are not well compatible with the structure and behaviour of living organisms (Glad and Ljung, 2000; Franklin et al., 2002). In addition to the already mentioned nonlinear characteristics of signal transmission blocks in the body, problems also include the circumstance that all biological processing structures are positive systems (i.e. their state variables can never be negative) and that the vertical translation from molecular to systems-level properties and vice-versa is difficult. To address the requirements of systems research in biomedicine we have developed a novel theory of nonlinear feedback loops that is compatible with basic physiological and biochemical knowledge (Dietrich and Boehm, 2006; Dietrich and Boehm, 2015). This theory is supported by specific methods of computer simulation that have been tailored to the requirements of life sciences.
At present, basically, three approaches to simulation are pursued, namely modelling in domain-specific environments (e.g. in the Systems Biology Markup Language [SBML] or solutions for System Dynamics such as Vensim, Stella, ithink or Powersim), toolkits in high-level computer algebra systems (e.g. Simulink for MATLAB) or custom simulation systems constructed bottom-up in universal programming languages (Neuber, 1989). The latter approach allows for higher flexibility and performance of the simulation, but it requires re-inventing the wheel, since for every simulation the elements of the processing structure must be programmed anew.
In the previous decades, our group has developed multiple mathematical models of biological control systems, especially in the field of endocrinology and metabolism. Examples include the hypothalamus-pituitary-thyroid (HPT) axis (Dietrich et al., 1997; Dietrich et al., 2004; Hoermann et al., 2013; Midgley et al., 2013; Hoermann et al., 2015; Berberich et al., 2018; Wolff et al., 2022), the hypothalamus-pituitary-adrenal (HPA) axis (Dietrich and Boehm, 2006) and insulin-glucose homeostasis (Dietrich et al., 2022). For a more detailed analysis of the HPT axis, we have developed a simulation program (SimThyr) that is based on our theory of nonlinear endocrine and metabolic feedback loops (Dietrich, 2002; Dietrich, 2017). For optimising future modelling efforts and to facilitate the rapid development targeting of other feedback loops, we decided to design a class library that supports the development of performant simulations and that is derived from the principles established for SimThyr.
1.2. Objectives
We, therefore, intended to develop a new approach that combines the advantages of the multiple avenues of modelling by providing a reusable class library for high-performance simulations.
The objectives of the project included the following tasks:
• To provide a standardised toolkit for the rapid development of simulation software;
• To support simulation in time domain and frequency domain;
• To provide certain dynamic elements that are typical for living organisms;
• To achieve high performance of the created software solutions in terms of simulation speed, reliability and memory efficiency;
• To achieve the scalability of the generated models by supporting different temporal resolutions;
• To support the drawing of block diagrams to screen windows;
• To foster dimensional analysis and vertical translation between a molecular and an organism-wide level.
2. Methods
2.1. Mathematical Theory
The theory of linear feedback loops is well covered in the classical systems theory (DiStefano, 1990) and only briefly mentioned here (Fig. 1). Linear control systems follow the superposition principle, which is defined by additivity

Figure 1. Linear feedback loops of zeroth order (A) and first order (B). x: set point or reference input; y: controlled variable; v: rate of change of controlled variable; yR: measured variable; z: load; e: error signal; u: controller output; G1: controller gain; G2: plant gain; α: gain of input signal in ASIA element; β: gain of signal deletion in ASIA element; ω: angular frequency
and homogeneity
The concept of linear time-invariant (LTI) systems is dealt with by a mature theoretical structure today (Röhler, 1973; Varju, 1977; Glad and Ljung, 2000; Franklin et al., 2002). Methods have been developed for the analysis of the steady-state behaviour of linear feedback loops, of transitional effects in the time and frequency domain, and in the form of state space description. However, the usefulness of the LTI theory for living systems is rather limited.
In the organism, another nonlinear class of feedback control systems is common. It is marked by saturable properties of both stimulating and inhibiting elements. For a large class of biological phenomena, stimulating subsystems can be modelled with the Monod equation (applied as Michaelis-Menten kinetics, receptor theory and the Langmuir adsorption model) as
where G denotes a maximum response (e.g., a secretory capacity or maximum enzyme activity under stimulated conditions) and D the magnitude of the input x that yields a half-maximum response.
Inhibiting subsystems are in most cases preferably represented as non-competitive inhibition with
where w and x denote stimulating and inhibiting input signals, respectively. Combining these nonlinear elements delivers a form of feedback loop that is well-grounded in physiological foundations (Fig. 2).

Figure 2. Nonlinear MiMe-NoCoDI loop comprising saturation (Michaelis-Menten-like, MiMe) kinetics and non-competitive inhibition (NoCoDI) elements (Dietrich and Boehm, 2015)
Although the question of steady-state prognosis has been solved for an important class of nonlinear feedback loops (Dietrich and Boehm, 2015), no consistent mathematical methodology is available for the analysis of their temporal behaviour. This limitation can be overcome with computer simulations. To enable their rapid and efficient implementation, the following class library was developed.
2.2. Implementation of a Class Library
Expanding the theoretical issues of Section 2.1, we have developed CyberUnits Bricks, a universal class library that allows for the implementation of simulation software for a large range of linear and nonlinear feedback loops, including the kind of feedback control systems mentioned.
CyberUnits Bricks library is available along with documentation as open-source software with a BSD licence from https://cyberunits.sourceforge.net or https://doi.org/10.5281/zenodo.1304179. Supplementary material including source code for the benchmark programs is hosted by Zenodo, operated by CERN, and available via https://doi.org/10.5281/zenodo.8317319.
The class library comprises five units. The Bricks unit provides basic functionality that is sufficient for the modelling of all common variants of linear feedback control systems. The Lifeblocks unit includes additional classes for the simulation of heterogeneous nonlinear systems, as they are typical for living organisms. The SystemsDiagram unit supports the generation of block diagrams corresponding to the model structure. It was intentionally separated from the Bricks and Lifeblocks units so as not to thwart the development of non-visual programs (e.g., for command line interfaces or server-site software). Functionality for the analysis of time series with fast Fourier transform is included in the SignalAnalysis unit. The plots unit supports the graphical analysis of processing structures with Bode plots.
Three of the five units are object-oriented and provide classes for simulation and modelling (Fig. 3). Basic classes for all purposes are available in the Bricks unit, and more specialised functionality for the simulation of living organisms in the LifeBlocks unit. Visual classes for drawing blocks diagrams are collected in the SystemsDiagram unit. The units Plots and SignalAnalysis contain procedural code for Bode plots and fast Fourier transform of time series.
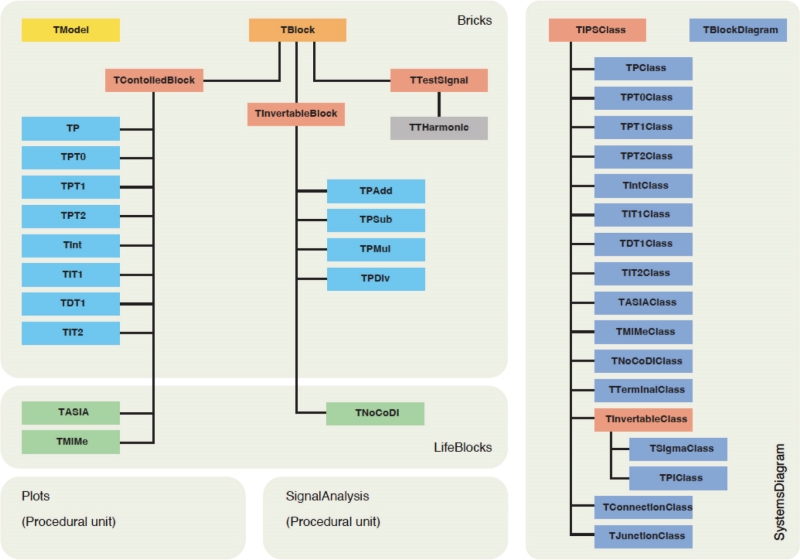
Figure 3. Class hierarchy of the CyberUnits Bricks library. See text and Table 1 for a more thorough explanation
A short summary of the available classes is provided in Table 1. Detailed information on the classes and units is provided in the documentation, which is available with the software, as described at the end of this article.
Table 1. Classes of the CyberUnits Bricks library
Class (for simulation) Units: Bricks and LifeBlocks |
Class (for block diagram) Unit: Systems Diagram |
Description |
TModel |
TIPSClass, TBlockDiagram |
Container class for blocks and fundamental properties of an information processing structure (IPS) |
TFR |
|
Representation of a frequency response as magnitude and phase |
TBlock |
|
Abstract block class |
TControlledBlock |
|
Abstract class for controlled blocks with input and output signals |
TInvertableBlock |
TInvertableClass |
Abstract class for “invertible” blocks with two opposing input signals |
TTHarmonic |
|
Representation for a harmonic test signal |
TP |
TPClass |
Class for a proportional element |
TPT0, TPT1, TPT2 |
TPT0Class, TPT1Class, TPT2Class |
Dead-time, first order and second order delay elements, respectively |
TInt |
TIntClass |
Class for an integrator |
TIT1, TIT2, TDT1 |
TIT1Class, TIT2Class, TDT1Class |
Complex elements with integrating, differential and delay components |
TPAdd, TPSub, TPMul, TPDiv |
TSigmaClass, TPiClass |
Invertible elements (adders, subtractors, multipliers and dividers, resp.) |
TASIA |
TASIAClass |
ASIA element (analog signal memory with intrinsic adjustment) |
TMiMe |
TMiMeClass |
Monod element (Michaelis-Menten or Langmuir kinetics) |
TNoCoDI |
TNoCoDIClass |
Non-competitive divisive inhibition |
|
TTerminalClass |
Draws terminal for external signal |
|
TConnectionClass |
Draws a connection between plots with arrow head and, if required, rounded corners |
|
TJunctionClass |
Draws a branching point |
The class library was primarily implemented in Object Pascal. The reasons for this decision included the fact that Object Pascal is type-safe, available for a plethora of platforms, holds a well-readable source code, supports multiple modern programming paradigms and generates native and fast machine code with contemporary compilers (Kamburelis, 2023). It was written with the Lazarus IDE for the Free Pascal compiler (Free Pascal Team, 1991-2021; Lazarus Team, 1993-2023). All non-graphical units of the class library were extended with conditionals to support Embarcadero Delphi as well. The minimum requirements for the Object Pascal version are Free Pascal 3.2.2 and Lazarus 2.2.6 or newer, or Delphi 11.3 or newer, respectively. Simplified versions for Swift and C++ were written with Swift 5.8.1 or C++20 with GNU extensions, respectively, in Xcode 14.3.1 on macOS 13.4.1. Scripts for S and Python were text files that were interpreted and executed by the corresponding environments (R version 4.2.3 for macOS and Python 3.9.6 with Clang 14.0.3 on Darwin, respectively).
2.3. Speed Comparison
In addition to the implementation in Object Pascal, a representative subset of the library was also implemented in additional programming languages, including S, Python, C++ and Swift in order to compare the simulation speed among versions in different languages. S (in the R implementation) and Python are interpreted languages that enjoy high popularity in biomedical research and computational biology. C++ and Swift are compiled languages that were developed with the goal of being robust and type-safe and generating optimised and therefore fast machine code. Despite certain language-specific adaptations, the fundamental structure of the class library is identical in all language versions.
Two simulation programs, one for a first-order feedback loop (Fig. 1B) and one for a MiMe-NoCoDI loop (Fig. 2), were written in S, Python, Swift, C++ and Object Pascal. The programs were linked to the above-mentioned Bricks and Lifeblocks class libraries written in the respective languages. As with the libraries, language-specific adaptations were used, but the basic structure of the program flow was identical in all four implementations (Fig. 4).

Figure 4. Exemplary algorithms for basic simulation of a 0th-order MiMe-NoCoDI loop (Fig. 2) and time measurements in Python, Swift and Object Pascal. See the online supplement for full source code in all evaluated programming languages
The simulations were implemented as command line interface (CLI) programs that were controlled via a Unix shell script. The shell script provided a loop mechanism that repeatedly invoked the simulation program with a systematic variation of the number of iteration steps (proportional to simulated time), ranging from 100 to 100,000 iterations with a step width of 50 iterations. Each step was repeated ten times so that benchmarks of 19,990 simulation runs were recorded for each programming language.
All simulation programs were executed on a MacBook Air Computer with Apple Silicon hardware (M2 processor, 8 cores, clock rate of up to 3.94 GHz, 24 GB of RAM) running macOS 13.4.1. During benchmarking, all non-essential services and background tasks were deactivated on the test machine. The software was either compiled for the AArch64 (A64) architecture or executed with an interpreter native for this instruction set.
2.4. Statistical Analysis
For a graphical representation of speed analysis, the mean values of all ten repetitions for each number of iterations were recorded and plotted. For statistical analysis, however, the data were evaluated in their original domain. Spearman correlations were used for the study of a potential relationship between iteration number and required time. For evaluating the correlations for different programming languages, models for pairs of two languages were compared under the assumption of independence with Fisher’s exact z-test and Zou’s confidence interval. The null hypothesis was that all correlations were equal. A p-value below an alpha-error threshold of 0.05 was regarded as significant.
To assess the relative contributions of the number of iterations and the programming language to the time of simulation a multivariable analysis was conducted, where a generalised linear model was fitted with an identity link function mapped to a gamma distribution.
All analyses were conducted with custom S scripts written for the statistical environment R (version 4.2.3 for macOS, R Core Team, 2023).
3. Results
3.1. Platform Availability
Thanks to the universal compatibility of modern Object Pascal compilers the class library is available for a large range of contemporary processor architectures (including x86-32, x86-64, PowerPC, ARM [AArch32 and AArch64]) and operating systems (e.g., MS Windows, macOS and several Linux variants). Major portions of the code have been implemented with compiler directives and conditionals to support both Free Pascal and Embarcadero Delphi. Visual classes have been written for the Lazarus Component Library (LCL) of the Lazarus IDE.
Test cases of the CyberUnits Bricks library and demo applications have been run on macOS, MS Windows and Raspbian Linux with Intel and ARM processors on 32-bit and 64-bit machines (Fig. 5).
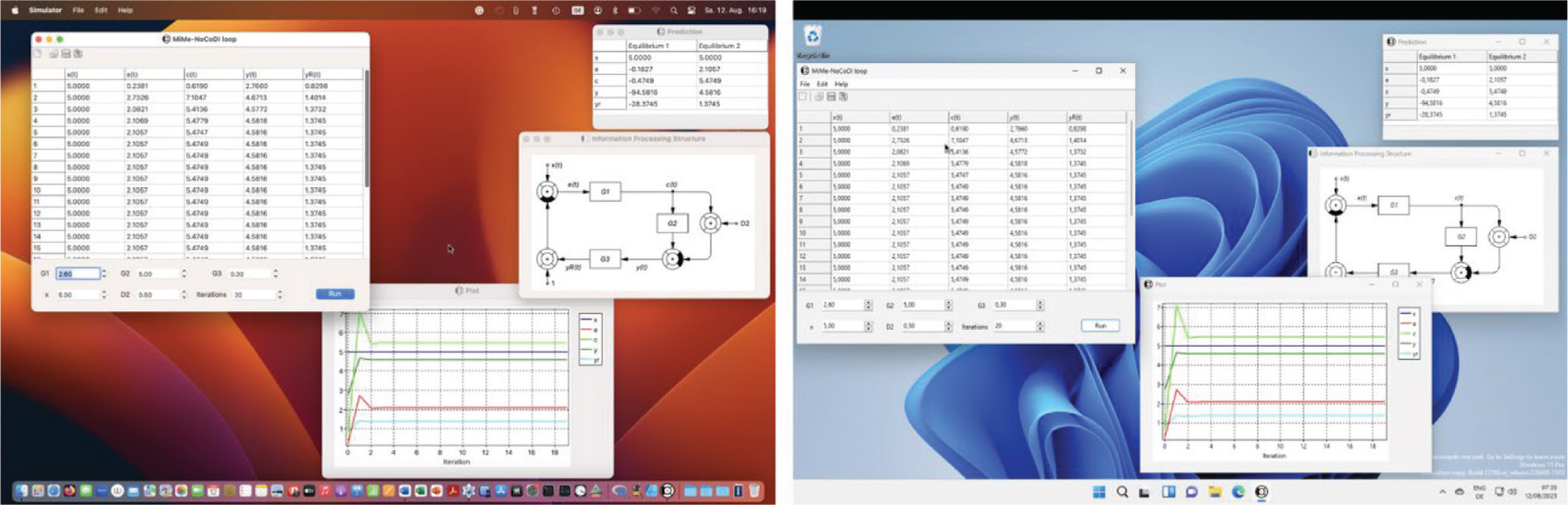
Figure 5. A simple simulator for a zeroth order MiMe-NoCoDI loop (see Fig. 2) written with CyberUnits Bricks on macOS 13.4 Ventura (Apple Silicon architecture, left) and Windows 11 (x86-64 architecture, right). The two implementations share the same source code, but run with native machine code and are optimized for the respective operating system as provided by the Lazarus Component Library (LCL) and the Free Pascal compiler. Shown are windows for time series in table form (top left of each screenshot) and graphical representation (bottom), the information processing structure as facilitated by the SystemsDiagram unit (right) and a steady-state prediction (top right, see Fig. 2 for a derivation of the equations). The simulated values rapidly approach the analytically predicted steady-state solution.
3.2. Simulation Speed
The speed of simulations was determined as the time needed for the completion of a simulation task (tc) with different numbers of iterations (i). The number of iterations translates to simulated time (ts) with
where δ represents the (uniform) step width of time-dependent simulation elements. Unlike ts, tc depends on the speed of the hardware and certain implementation details of the programming language and the operating system.
Irrespective of the number of iterations, Object Pascal yields the fastest and S the slowest programs. C++, Swift and Python are in between. The results for the two simulated feedback loops and the four tested programming languages are shown in Fig 6. For all languages, the required time significantly correlated with the number of iterations (p < 2.2e–16 for all relationships). Additionally, the correlations were different for all languages (p < 1e–4 and Zou’s 95% confidence interval not including 0).
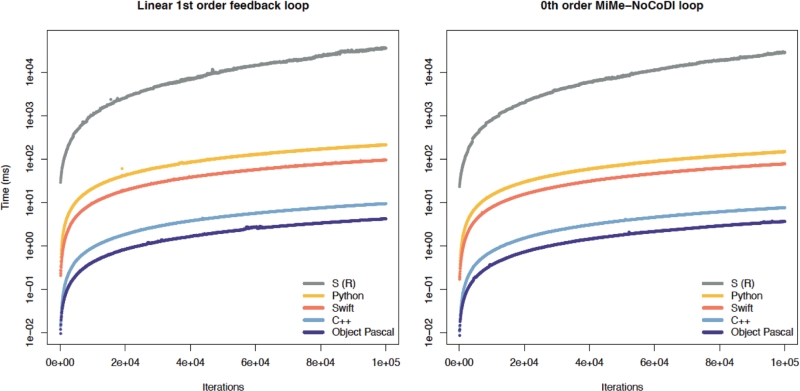
Figure 6. Simulation speed with implementations in different programming languages. Shown is the time needed to simulate a first-order linear feedback loop (A) and a zeroth-order MiMe-NoCoDI loop (B) dependent on the number of iterations (proportional to simulated time, abscissa) based on implementations of the simulation program and the CyberUnits Bricks library in S, Python, Swift, C++ and Object Pascal. The ordinate is logarithmically scaled and represents the mean of 10 replications of each number of iteration steps
In the models of multivariable regression analysis, the number of iterations and the programming languages are independent predictors of the time needed for simulation (Tables 2 and 3), with the exception of the pair of Swift and C++, which was not different. A variance inflation factor (VIF) of about one denotes virtually absent multicollinearity.
Table 2. Models of multivariable regression analysis with respect to time needed for simulation of a first-order linear feedback loop (in milliseconds). B: Beta coefficient; SE: standard error; Wald: Wald’s chi-squared statistics; d.f.: degrees of freedom; p-value: alpha error; VIF: variance inflation factor
|
B |
SE |
Wald |
d.f. |
p-value |
VIF |
Reference: Object Pascal |
||||||
Iterations |
4.371e–5 |
1.844e–7 |
56192.6 |
1 |
< 2e–16 |
1.0048 |
S |
1.301e+4 |
52.790 |
60756.5 |
1 |
< 2e–16 |
1.0048 |
Python |
1.028e+2 |
0.4259 |
58264.9 |
1 |
< 2e–16 |
|
Swift |
44.110 |
0.1878 |
55159.8 |
1 |
< 2e–16 |
|
C++ |
44.110 |
0.1878 |
55159.8 |
1 |
< 2e–16 |
|
Reference: S |
||||||
Iterations |
4.371e–5 |
1.844e–7 |
56192.6 |
1 |
< 2e–16 |
1.0048 |
Python |
–1.291e+4 |
52.790 |
59796.3 |
1 |
< 2e–16 |
1.0048 |
Swift |
–1.297e+4 |
52.790 |
60344.5 |
1 |
< 2e–16 |
|
C++ |
–1.297e+4 |
52.790 |
3.8 |
1 |
< 2e–16 |
|
Reference: Python |
||||||
Iterations |
4.371e–5 |
1.844e–7 |
56192.6 |
1 |
< 2e–16 |
1.0048 |
Swift |
–58.700 |
0.4653 |
15914.3 |
1 |
< 2e–16 |
1.0048 |
C++ |
–58.700 |
0.4653 |
15914.3 |
1 |
< 2e–16 |
|
Reference: Swift |
||||||
Iterations |
4.371e–5 |
1.844e–7 |
56192.6 |
1 |
< 2e–16 |
1.0048 |
C++ |
6.421e–11 |
0.2650 |
5.9e–20 |
1 |
n. s. |
1.0048 |
Table 3. Models of multivariable regression analysis with respect to the time needed for simulation of a zeroth order MiMe-NoCoDI loop (in milliseconds). See Table 2 for abbreviations
|
B |
SE |
Wald |
d.f. |
p-value |
VIF |
Reference: Object Pascal |
||||||
Iterations |
3.776e–5 |
1.585e–7 |
56789.6 |
1 |
< 2e–16 |
1.0056 |
S |
1.027e+4 |
41.410 |
61459.1 |
1 |
< 2e–16 |
1.0056 |
Python |
70.720 |
0.2929 |
58315.6 |
1 |
< 2e–16 |
|
Swift |
35.700 |
0.1516 |
55445.6 |
1 |
< 2e–16 |
|
C++ |
35.700 |
0.1516 |
55455.6 |
1 |
< 2e–16 |
|
Reference: S |
||||||
Iterations |
3.776e–5 |
1.585e–7 |
56789.6 |
1 |
< 2e–16 |
1.0056 |
Python |
–1.019e+4 |
41.410 |
60612.1 |
1 |
< 2e–16 |
1.0056 |
Swift |
–1.023e+4 |
41.410 |
61031.5 |
1 |
< 2e–16 |
|
C++ |
–1.023e+4 |
41.410 |
2.9 |
1 |
< 2e–16 |
|
Reference: Python |
||||||
Iterations |
3.776e–5 |
1.585e–7 |
56789.6 |
1 |
< 2e–16 |
1.0056 |
Swift |
–35.020 |
0.3296 |
11290.0 |
1 |
< 2e–16 |
1.0056 |
C++ |
–35.020 |
0.3296 |
11290.0 |
1 |
< 2e–16 |
|
Reference: Swift |
||||||
Iterations |
3.776e–5 |
1.585e–7 |
56789.6 |
3.776e–5 |
1.585e–7 |
1.0056 |
C++ |
–7.334e–11 |
0.2142 |
1.2e–19 |
1 |
n. s. |
1.0056 |
4. Discussion
With CyberUnits Bricks a versatile, cross-platform class library is available that supports modelling and simulation in life sciences. Its focus is on the simulation of feedback loops and similar motifs in systems biology.
We could provide evidence that the implementation in Object Pascal delivers a high performance of the generated modelling applications. Our results fit earlier observations which demonstrated that Pascal, the older non-object-oriented variant of the language, delivered the fastest implementation of several algorithms (QuickSort, Hailstone, sieve of Eratosthenes) compared to other programming languages, including C, C++, Go, Fortran, Rust and Python (Pereira et al., 2021). Additionally, Pascal was among the three Pareto optimal languages for the combination of time, memory usage and energy consumption of the hardware among a total of 27 programming languages (Pereira et al., 2021).
It is not the intention of the class library to replace existing products for modelling and simulation. Along these lines, it was not envisioned to offer an “out-of-the-box” solution by delivering directly usable simulation programs. The objective was, rather, to provide a versatile infrastructure for the creation of high-performance simulation solutions for complex systems in life sciences. Additionally, the availability of the class library helps to avoid reinventing the wheel by providing versatile and reusable building bricks of biological information processing structures for a range of platforms. Due to the implementation in a universal programming language, it can also lay the foundation for the making of stand-alone applications.
The class library has been successfully used in a simulation program for insulin-glucose homeostasis (Dietrich & Boehm, 2021; Dietrich et al., 2022) and is currently being employed to provide the basis for the next major version of a simulator of thyroid homeostasis for widespread use (SimThyr, Dietrich, 2017). When used with integrated development environments for rapid application development (RAD), such as Embarcadero Delphi or Lazarus, CyberUnits Bricks opens the way to the fast development of simulation programs. On this basis, an experienced developer can write a finished program for a simple feedback loop in less than 30 minutes.
On the other hand, the development of a simulation program requires some knowledge of programming. Therefore, CyberUnits Bricks is less suited for basic educational projects or the occasional use of simulation methods by unsophisticated persons without any background in software development. Its strengths are in versatility and performance.
5. Conclusion
CyberUnits Bricks is a universal class library that supports the development of computer simulations in life sciences with a focus on feedback loops and other control motifs in systems biology. Its main fork is developed in Object Pascal, thereby ensuring the high performance of the created software.
Future applications of the class library may target a large area of biomedical information processing structures and network motifs. This will require the support of additional signal transmission blocks, including Hill kinetics, and competitive and uncompetitive inhibition.
Currently, the Bricks library is being extended to fully support Embarcadero Delphi and RAD Studio as well. Thereby, developers and scientists will have more freedom of choice regarding the integrated development environment, and it will be possible to target a larger spectrum of platforms, ranging from smartphones to supercomputers.
Thanks to the high speed and the broad hardware support provided by the Object Pascal language, solutions built with CyberUnits Bricks may also find their way into devices of medical technology, e.g. insulin pumps with advanced hybrid closed-loop (AHCL) and automated insulin delivery (AID) algorithms. This step will considerably extend the scope of the class library from simulation to medical engineering, but it will also require the inclusion of extensive safety measures, as provided e.g. by the IEC 62304 standard and NASA’s Power of 10 rules (Holzmann, 2006).
6. Acknowledgements
This report is an extended version of a keynote lecture and a software presentation provided by the first author at the International Pascal Congress, held from July 3rd to 7th, 2023, in Salamanca.
7. References
Alon, U. (2007). Network motifs: theory and experimental approaches. Nature Reviews Genetics, 8, )450-61. https://doi.org/10.1038/nrg2102
Alon, U. (2020). An Introduction to Systems Biology. CRC Press.
Berberich, J., Dietrich, J. W., Hoermann, R., & Mueller, M. A. (2018). Mathematical Modeling of the Pituitary-Thyroid Feedback Loop: Role of a TSH-T-3-Shunt and Sensitivity Analysis. Front Endocrinol (Lausanne), 9. https://doi.org/10.3389/fendo.2018.00091
Cruz-Loya, M., Chu, B. B., Jonklaas, J., Schneider, D. F., & DiStefano, J., 3rd (2022). Optimized Replacement T4 and T4+T3 Dosing in Male and Female Hypothyroid Patients With Different BMIs Using a Personalized Mechanistic Model of Thyroid Hormone Regulation Dynamics. Frontiers in endocrinology, 13, 888429. https://doi.org/10.3389/fendo.2022.888429
Curcio, L., D'Orsi, L., Cibella, F., Wagnert-Avraham, L., Nachman, D., & De Gaetano, A. (2020). A Simple Cardiovascu-lar Model for the Study of Hemorrhagic Shock. Computational and mathematical methods in medicine, 2020, 7936895. https://doi.org/10.1155/2020/7936895
Dietrich, J. W. (2002). Der Hypophysen-Schilddrüsen-Regelkreis. Entwicklung und klinische Anwendung eines nichtlinearen Modells (Vol. 2). Logos-Verlag.
Dietrich, J. W. (2017). SimThyr. Report No. RRID:SCR_014351, (Zenodo, 2017).
Dietrich, J. W., & Boehm, B. O. (2006). Equilibrium behaviour of feedback-coupled physiological saturation kinetics. In Trappl R. (ed.), Cybernetics and Systems 2006 (Vol. 1, pp. 269-274). Austrian Society for Cybernetic Studies.
Dietrich, J. W. & Boehm, B. O. (2015). Die MiMe-NoCoDI-Plattform: Ein Ansatz für die Modellierung biologischer Regelkreise. German Med. Sci. DocAbstr. 284. https://doi.org/10.3205/15gmds058
Dietrich, J. W. & Boehm, B. O. (2021). SimulaBeta. Report No. RRID: SCR_021900, (Zenodo, 2021).
Dietrich, J. W., Dasgupta, R., Anoop, S., Jebasingh, F., Kurian, M. E., Inbakumari, M., Boehm, B. O., & Thomas, N. (2022). SPINA Carb: a simple mathematical model supporting fast in-vivo estimation of insulin sensitivity and be-ta cell function. Scientific reports, 12(1), 17659. https://doi.org/10.1038/s41598-022-22531-3
Dietrich, J. W., Landgrafe-Mende, G., Wiora, E., Chatzitomaris, A., Klein, H. H., Midgley, J. E., & Hoermann, R. (2016). Calculated Parameters of Thyroid Homeostasis: Emerging Tools for Differential Diagnosis and Clinical Research. Frontiers in endocrinology, 7, 57. https://doi.org/10.3389/fendo.2016.00057
Dietrich, J. W., Mitzdorf, U., Weitkunat, R., & Pickardt, C. R. (1997). The pituitary-thyroid feedback control: stability and oscillations in a new nonlinear model. Journal of Endocrinological Investigation, 20 (Suppl. to no. 5), 100.
Dietrich, J. W., Tesche A., Pickard C. R. and Mitzdorf U. (2004). Thyrotropic feedback control: Evidence for an addition-al ultrashort feedback loop from fractal analysis. Cybernetics and Systems, 35(4), 315-331. https://doi.org/10.1080/01969720490443354
DiStefano, J. (1990). Schaum’s Outline of Feedback and Control System. McGraw-Hill.
Franklin, G. F., Powell, J. D., Emami-Naeini, A. (2002). Feedback Control of Dynamic Systems. Prentice Hall.
Free Pascal Team. (1993-2023). Free Pascal: A 32-, 64- and 16-bit professional Pascal compiler. Report No. RRID: SCR_014360, (Fair-fax, VA, 1993-2023).
Glad, T., Ljung, L. (2000). Control Theory. Taylor & Francis.
Han, S. X., Eisenberg, M., Larsen, P. R., & DiStefano, J., 3rd (2016). THYROSIM App for Education and Research Pre-dicts Potential Health Risks of Over-the-Counter Thyroid Supplements. Thyroid: official journal of the American Thyroid Association, 26(4), 489-498. https://doi.org/10.1089/thy.2015.0373
Head, R. J., Lumbers, E. R., Jarrott, B., Tretter, F., Smith, G., Pringle, K. G., Islam, S., & Martin, J. H. (2022). Systems analysis shows that thermodynamic physiological and pharmacological fundamentals drive COVID-19 and re-sponse to treatment. Pharmacology research & perspectives, 10(1), e00922. https://doi.org/10.1002/prp2.922
Hoermann, R., Midgley, J. E. M., Larisch, R., & Dietrich, J. W. (2013). Is pituitary TSH an adequate measure of thyroid hormone-controlled homoeostasis during thyroxine treatment? European Journal of Endocrinology, 168(2), 271-280. https://doi.org/10.1530/EJE-12-0819
Hoermann, R., Midgley, J. E., Larisch, R., & Dietrich, J. W. (2015). Homeostatic Control of the Thyroid-Pituitary Axis: Perspectives for Diagnosis and Treatment. Front Endocrinol (Lausanne), 6, 177. https://doi.org/10.3389/fendo.2015.00177
Hoermann, R., Pekker, M. J., Midgley, J. E. M., Larisch, R., & Dietrich, J. W. (2022). Principles of Endocrine Regulation: Reconciling Tensions Between Robustness in Performance and Adaptation to Change. Frontiers in endocrinology, 13, 825107. https://doi.org/10.3389/fendo.2022.825107
Holzmann, G. (2006). The Power of 10: Rules for Developing Safety-Critical Code. IEEE Computer. 39(6), 95-9. https://doi.org/10.1109%2FMC.2006.212
Kamburelis, M. (2023). Why use Pascal? https://castle-engine.io/why_pascal
Kolar-Anić, L., Čupić, Ž., Maćešić, S., Ivanović-Šašić, A., & Dietrich, J. W. (2023). Modelling of the thyroid hormone synthesis as a part of nonlinear reaction mechanism with feedback. Computers in biology and medicine, 160, 106980. https://doi.org/10.1016/j.compbiomed.2023.106980
Lazarus Team. (1993-2001). Lazarus: The professional Free Pascal RAD IDE. Report No. RRID: SCR_014362, (Fairfax, VA, 1993-2021).
McEwen B. S. (1998). Stress, adaptation, and disease. Allostasis and allostatic load. Annals of the New York Academy of Sciences, 840, 33-44. https://doi.org/10.1111/j.1749-6632.1998.tb09546.x
McEwen, B. S., & Stellar, E. (1993). Stress and the individual. Mechanisms leading to disease. Archives of internal medicine, 153(18), 2093-2101. https://doi.org/10.1001/archinte.1993.00410180039004
Midgley, J. E. M., Hoermann, R., Larisch, R., & Dietrich, J. W. (2013). Physiological states and functional relation be-tween thyrotropin and free thyroxine in thyroid health and disease: in vivo and in silico data suggest a hierarchical model. J Clin Pathol, 66(4), 335-342. https://doi.org/10.1136/jclinpath-2012-201213
Neuber, H. (1989). Simulation von Regelkreisen auf Personal Computern in Pascal und Fortran 77. IWT.
Pereira, R., Couto, M., Ribeiro, F., Rua, R., Cunha, J., Fernandes, J. P., & Saraiva, J. (2021). Ranking programming lan-guages by energy efficiency. Science of Computer Programming, 205, 102609. https://doi.org/10.1016/j.scico.2021.102609
Pompa, M., Panunzi, S., Borri, A., & De Gaetano, A. (2021). A comparison among three maximal mathematical models of the glucose-insulin system. PloS one, 16(9), e0257789. https://doi.org/10.1371/journal.pone.0257789
R Core Team. (2023). R: A Language and Environment for Statistical Computing. Report No. RRID:SCR_001905, (R Foundation for Statistical Computing, Vienna, Austria, 2018).
Röhler, R. (1973). Biologische Kybernetik - Regelungsvorgänge in Organismen. Vieweg+Teubner Verlag. https://doi.org/10.1007/978-3-322-94729-1
Schulkin, J., & Sterling, P. (2019). Allostasis: A Brain-Centered, Predictive Mode of Physiological Regulation. Trends in neurosciences, 42(10), 740-752. https://doi.org/10.1016/j.tins.2019.07.010
Tretter F. (2018). From mind to molecules and back to mind-Metatheoretical limits and options for systems neuropsychiatry. Chaos, 28(10), 106325. https://doi.org/10.1063/1.5040174
Tretter, F., Wolkenhauer, O., Meyer-Hermann, M., Dietrich, J. W., Green, S., Marcum, J., & Weckwerth, W. (2021). The Quest for System-Theoretical Medicine in the COVID-19 Era. Frontiers in medicine, 8, 640974. https://doi.org/10.3389/fmed.2021.640974
Tretter, F., Peters, E. M. J., Sturmberg, J., Bennett, J., Voit, E., Dietrich, J. W., Smith, G., Weckwerth, W., Grossman, Z., Wolkenhauer, O., & Marcum, J. A. (2023). Perspectives of (/memorandum for) systems thinking on COVID-19 pandemic and pathology. Journal of evaluation in clinical practice, 29(3), 415-429. https://doi.org/10.1111/jep.13772
Varjú, D. (1977). Systemtheorie für Biologen und Mediziner. Springer-Verlag,
Wolff, T. M., Veil, C., Dietrich, J. W., & Muller, M. A. (2022). Mathematical modeling and simulation of thyroid homeo-stasis: Implications for the Allan-Herndon-Dudley syndrome. Front Endocrinol (Lausanne), 13, 882788. https://doi.org/10.3389/fendo.2022.882788